Description: Participants: Interactions 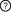 This is a sequential switch. The interactions in this switch take place in a defined order. The order is defined by numbers to the left of each interaction. More than one interaction can occur in each step, these interactions occur independently but are necessary for future step This is a sequential switch. The interactions in this switch take place in a defined order. The order is defined by numbers to the left of each interaction. More than one interaction can occur in each step, these interactions occur independently but are necessary for future step1Interaction #1 CKS1B - SKP2 Interfaces (1) Cyclin-dependent kinase regulatory subunit in Cyclin-dependent kinases regulatory subunit 1 (CKS1B) (2) Leucine rich repeat (205-229) and (230-254) and (255-284) and (309-334) and (335-359) in S-phase kinase-associated protein 2 (SKP2) Additional Information Affinity : 0.006 µM Structural information: 2ASS 1Interaction #2 CDKN1B - CDK2 Interfaces (3) MOD_CDK_1 motif (184190) in Cyclin-dependent kinase inhibitor 1B (CDKN1B) (4) Protein kinase domain (4-286) in Cyclin-dependent kinase 2 (CDK2) 2Interaction #3 CDKN1B - Interaction #1 CKS1B - SKP2 Requires Interaction #1 CKS1B - SKP2 Requires Interaction #2 CDKN1B - CDK2 Interfaces (5) DEG_SCF_SKP2-CKS1_1 motif (183190) in Cyclin-dependent kinase inhibitor 1B (CDKN1B) (6) Interaction #1 CKS1B - SKP2 Interaction Regulation PTM-dependent Induction (Phosphorylation of T187 on Cyclin-dependent kinase inhibitor 1B (CDKN1B)) of the Cyclin-dependent kinase inhibitor 1B (CDKN1B) DEG_SCF_SKP2-CKS1_1 motif - interaction Regulatory Enzymes for switch Modifying enzymes for residue: T187: Cyclin-dependent kinase 2 (CDK2) Additional Information Affinity : 7 µM Structural information: 2AST Context Switch localisation Curated: nucleus ELM curated: nucleus, cytosol, cycloplasmic cyclin-dependent protein kinase holoenzyme complex, SCF ubiquitin ligase complex Switch cell cycle phase Curated: mitotic S phase Switch pathways Curated: REACTOME:2676436, Cell cycle References (1) Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. See also Other switches involving participants |
|
Please send any suggestions/comments to: switches@elm.eu.org

