Switch types and subtypes
- Binary on-off switch
- Specificity switch
- Cumulative switch
- Avidity-sensing switch
- Pre-assembly switch
Binary switch
Simple binary switches shift between two distinct functional states of a motif. This can be either an "on" and "off" state, or two states with different affinities for a single binding partner. Based on the mechanism that regulates switching between the two states, different subtypes were defined.
Altered physicochemical compatibility
PTM of a residue in a motif or in its flanking regions alters the physicochemical and/or structural compatibility of the motif with its binding partner. This can either induce or enhance an interaction, or result in inhibition or even abrogation of an interaction.
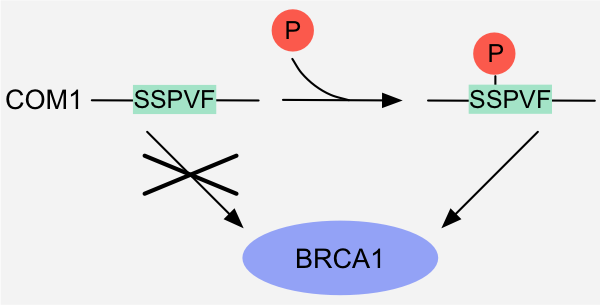
Figure 1: Binary switch - Altered physicochemical compatibility: binding of the BRCT-binding motif in the DNA endonuclease COM1 to the tandem BRCT
domains of BRCA1 is induced by phosphorylation of the serine residue in the motif 1.
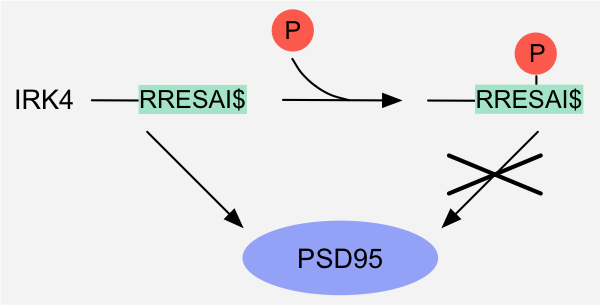
Figure 2: Binary switch - Altered physicochemical compatibility: binding of the PDZ-binding motif in the IRK4 potassium channel to the PDZ domain of DLG4/PSD95 is
inhibited by phosphorylation of the serine residue in the motif1.
Allostery
The binding properties of a motif or a motif-binding domain are modulated indirectly by allosteric effects resulting from PTM or effector binding at a site that is distinct from the actual interaction interface.
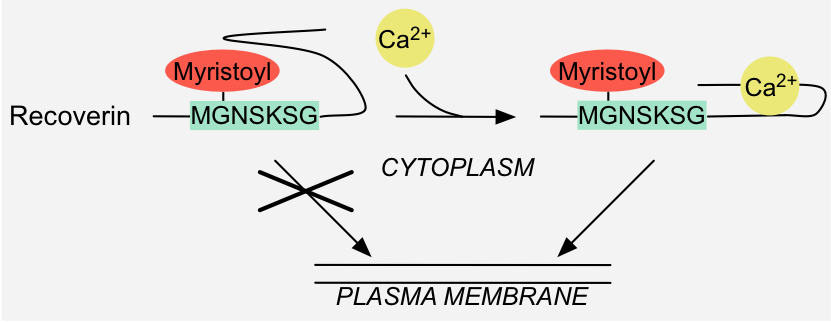
Figure 3: Binary switch - Allostery: binding of calcium to Recoverin results in conformational changes that make the covalently attached myristoyl moiety
accessible for recruitment to the plasma membrane 1.
Pre-translational switch
Pre-translational mechanisms such as alternative splicing, alternative promoter-usage and/or RNA editing result in inclusion or removal of exons that contain an entire or partial motif.
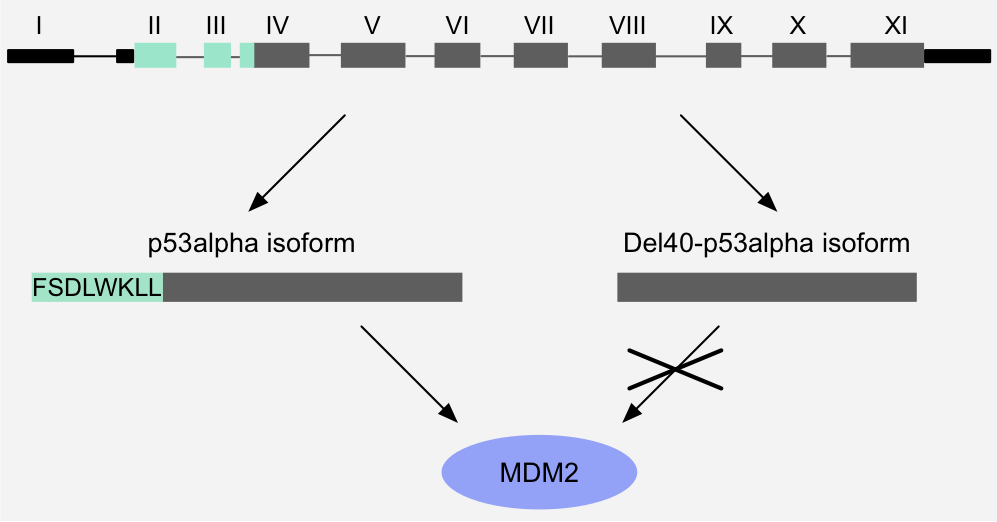
Figure 4: Binary switch - Pre-translational switch: alternative promoter usage and alternative splicing removes the MDM2-binding motif of p53, thereby abrogating binding to MDM2. As a result, this p53 isoform is resistant to MDM2-mediated degradation
and shows a longer half-life1,2.
Specificity switch
Specificity switches on overlapping or adjacent, mutually exclusive interaction interfaces are characterised by distinct "on" states, allowing regulated exchange of distinct binding partners. Different subtypes were defined based on the mechanism that regulates switching of binding partners.
Competitive binding
Competitive binding of multiple binding partners to overlapping or adjacent, mutually exclusive interaction interfaces depends on local target protein abundance, which can be regulated by changing the expression level or subcellular localisation of the competitors, or by scaffolding.
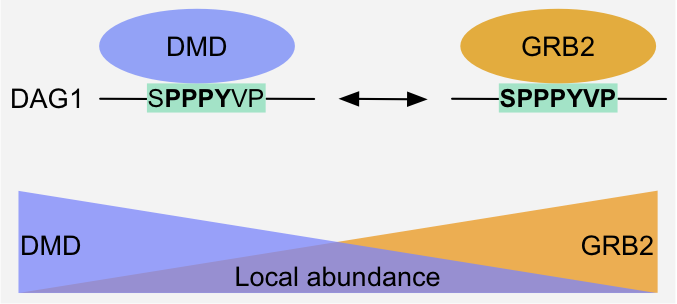
Figure 8: Specificity switch - Competition: Dystrophin and Grb2 compete for the overlapping WW-binding and SH3-binding motifs (in bold), respectively, of Dystroglycan.
The outcome of the competition depends on the local abundance of the competitors, which can be modulated by changing their expression level or localization and by
scaffolding1,2.
Altered binding specificity
The balance of the competition for overlapping or adjacent, mutually exclusive interaction interfaces is tipped in favor of one of the interactors by PTM-dependent modulation of the intrinsic affinity of a binding region. Multiple, successive PTMs allow sequential switching of different binding partners in an ordered manner by step-wise alteration of binding specificity.
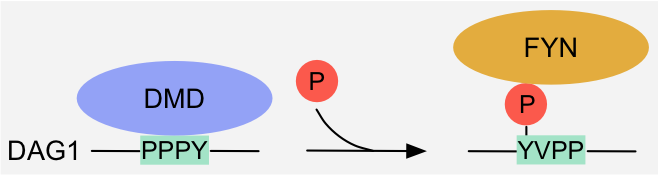
Figure 5: Specificity switch - Altered binding specificity: phosphorylation of the tyrosine residue in the overlapping WW- and SH2-binding motifs in
Dystroglycan switches the specificity from the WW domain protein Dystrophin to the SH2 protein Fyn kinase1.
Motif hiding
Motif hiding occurs when there is a large difference in intrinsic affinity of overlapping or adjacent motifs for their respective binding partners, or a large difference in the local abundance of these partners. Binding of an effector to one motif sterically masks the overlapping or adjacent motif, thereby precluding it from binding. Binding of the masking molecule can be PTM-dependent or -independent.
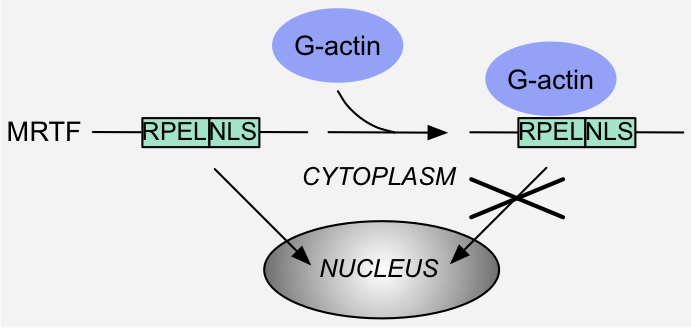
Figure 6: Specificity switch - Motif hiding: binding of monomeric G actin to the RPEL motifs in MRTF hides the Nuclear Localization Signal (NLS) and precludes
interaction with the import machinery, thereby excluding MRTF from the nucleus 1.
Domain hiding
A domain can be sterically masked by binding of an effector when there is a large difference in intrinsic affinity of the domain for different binding partners, or a large difference in the local abundance of these partners, thereby precluding further interactions of the domain. Binding of the masking molecule can be PTM-dependent or -independent.
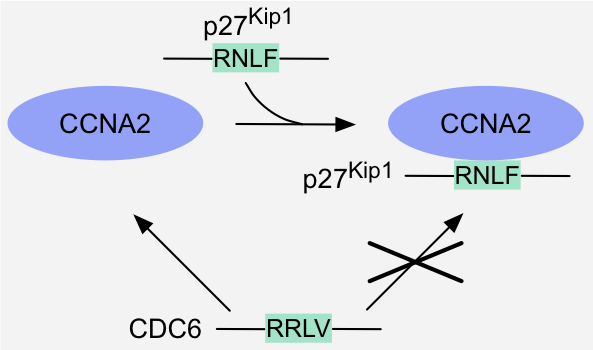
Figure 7: Specificity switch - Domain hiding: binding of the cyclin-Cdk inhibitor p27Kip1 to a cyclin-Cdk complex hides the binding site for
the cyclin-binding motifs present in cyclin-Cdk substrates like Cdc6, and thereby prevents recruitment of these substrates to the complex1.
Cumulative switch
Regulation of an interaction by a cumulative switching mechanism is mediated by multisite modification.
Rheostatic switch
Rheostatic switches gradually alter the affinity of a motif for a single binding partner by addition of multiple PTMs that additively contribute to this modulation. Additional modifications can either strengthen or weaken an interaction.
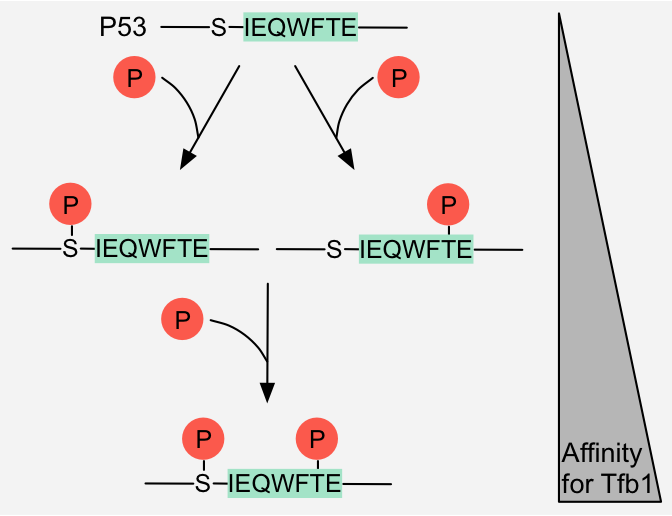
Figure 9: Cumulative switch - Rheostatic switch: Phosphorylation of a serine and threonine residue respectively flanking and in the Tfb1-binding motif in p53 additively
increases the affinity of p53 for Tfb1 1.
Avidity-sensing switch
Multiple low-affinity interactions give rise to high-avidity interactions that have increased binding strength, with more than additive affinity.
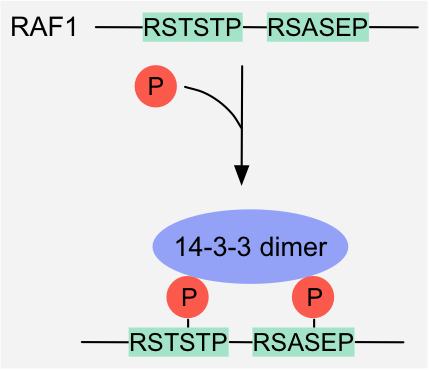
Figure 10: Avidity-sensing switch: Phosphorylation of two 14-3-3-binding motifs in Raf1 induces high-avidity binding to 14-3-3 dimers
1.
Pre-assembly switch
Pre-assembly switches require prior formation of a complex as a prerequisite for a motif-mediated interaction to occur.
Composite binding site formation
The formation of a complex results in the generation of a continuous motif-binding site that spans more than one component of this complex. Neither complex subunit on its own contains a functional binding domain for the motif, and interaction of the motif only occurs in the context of the active, fully assembled complex.
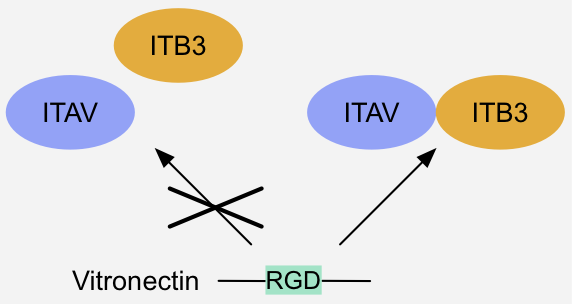
Figure 11: Pre-assembly switch - Composite binding site formation: Binding of integrin-receptor ligands like Vitronectin requires pre-assembly of a heterodimeric
integrin alpha-beta complex, since the integrin alpha and beta subunit together form a composite binding site for the ligand's RGD motif 1 .
NOTE: It should be taken into consideration that the examples curated in the database are regarded in isolation, however in a biological context these definitions partially overlap and different mechanisms are combined to mediate complex regulatory functions.