switches.ELM help page
- switches.ELM User Manual
- switches.ELM Data Content
- switches.ELM Downloads and Usage Policy
- switches.ELM User Data Submission
switches.ELM User Manual
Searching switches.ELM
The switches.ELM database can be searched in two different ways: browsing or key word-based searching.
Browse switches.ELM
The switches.ELM database can be browsed by different categories:
- Switch: lists all switches curated in the database.
- Protein: lists all proteins curated in the database.
- Motif class: lists all ELM motif classes curated in the database.
- Switch type: lists all switches curated in the database by their type and subtype.
Search switches.ELM
The switches.ELM database can be searched by search terms entered by the user. All the information that is present in the database, as described in the switches.ELM content section above, is used for term-based searching of the database. This means that searches can be done using the name of a protein, or its gene name or alternative names listed in UniProtKB, but also by PTM type or gene ontology terms that are associated to a protein, or by looking for species-specific proteins. The search results are returned as a list of switches, classified according to the data that matched to the query, which is mentioned at the top of each list.
Search output page / Graphical representation
Each switch instance in the switches.ELM database can be viewed in a visual output that gives a clear and detailed overview of the switching mechanism (Figure 1A). Hovering over switch types and subtypes or sequence features shows their definition or a description, respectively, while clicking on annotated data and features allows navigating to external resources of the data for more detailed information, or to publications providing evidence for the data. Schematic visualisation of the protein architecture shows the motif switch instance in the context of the whole protein (Figure 1B).

Figure 1A: Visual output of switch instances in the switches.ELM database: (Left) Data curated for a switch instance in the switches.ELM database. (1) Type and subtype, text description and graphical presentation of the switch instance. (2) List of interaction participants. (3) List of interactions constituting the switch instance. (3a) Interactions named by its two participants. If interactions occur in a specific order, they are numbered according to their position in the sequence of binding events. (3b) List of interactions that are mutually exclusive with this interactions, or, alternatively, that are required for this interaction to occur. (3c) List of binding interfaces that mediate the interaction between the two participants. (3d) Description for how an interaction is regulated. (3e) Additional information, including kinetic and structural information, when available. (4) List of relevant publications describing experimental data for the switch instance. (5) List of other switch instances involving any of the participants or interaction interfaces. (Right) Alignment and features for the region of interest. (6) Protein architecture (See figure 1B). (7) Toggle categories of information. (8) Expandable alignment of the region of interest. (9) Validated motifs and PTMs that participate in a curated switch instance. (10) Validated motifs in this region that are curated in ELM and predicted SLiMs based on the regular expressions defined for ELM motif classes. (11) Validated modification sites. If available, experimental mutated sites are shown. (12) Structural information for this region of the protein. (13) Information on protein isoform-specific expression of this region. (14) Single Nucleotide Polymorphisms (SNPs) found in this region. (15) Additional sequence features. (16) Disorder scores for this region calculated by IUPred.
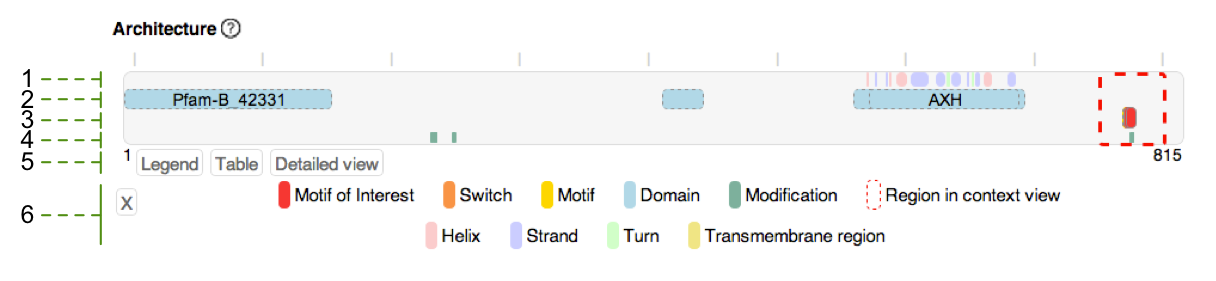
Figure 1B: Visualisation of the protein architecture: (1) Secondary structure elements. (2) Protein domains. (3) Experimentally validated SLiMs (from ELM) and switches (from switches.ELM), with the motif of interest highlighted in red. (4) Modification sites. (5) Buttons to show figure legend, information table (containing name and position of the sequence features), or a detailed view of the whole protein sequence. (6) Figure legend showing the colour code of the sequence features annotated for this protein. Clicking the 'Hide' button will hide the legend.
Protein analysis
Submit motif of interest
Using any of the identifiers in UniProtKB, a protein of interest can be analysed for possible motif-based switching mechanisms. If the search term is too general, a list of matching proteins is returned, from which the user can select the protein to be analysed. Next, the sequence of the protein is returned. In this sequence, the user can highlight the motif of interest, either by typing the motif sequence in the 'Highlight motif' box, or by directly clicking the motif residues in the sequence. Clicking the submit button will then start the analysis of the motif indicated by the user.
Analysis output page / Graphical representation
The results of the analysis of a user-submitted motif is shown in a graphical output similar to that used to visualise switch instances in the database (Figure 2).

Figure 2: Visual output of the switches.ELM prediction tool: (Left) Description of putative switches regulating a user-submitted motif. (1) General information, including the name and species of the query protein, the sequence of the query motif, possible matches to known ELM motif classes, and curated switches involving this protein. (2) Select buttons to view either a list of putative switches for the user-submitted motif, links to publications providing experimental evidence (for curated switches involving this protein, mutagenesis data available for this protein region, and structural information for the protein linked to PDB), or warnings concerning the structural context of the motif. (3) List of putative switches regulating the user-submitted motif, including available experimental data for modification sites that might be involved. Hovering over the question marks will show switch type definitions. (Right) Alignment and features for the region of interest. Data extracted from external resources for the aligned protein region is identical to those shown in the visual output of a curated switch instance (See figure 1A).
switches.ELM Data Content
Biomolecule data
Protein data
IDENTIFIERS: Proteins are identified by different identifiers collected from UniProtKB/Swiss-Prot: the primary accession number (e.g. P04637), secondary accession numbers (e.g. Q15086), entry name (e.g. P53_HUMAN), recommended name (e.g. Cellular tumor antigen p53), alternative name(s) (e.g. Tumor suppressor p53) and gene name (e.g. TP53) and synonym(s) (e.g. P53). Proteins are linked to their UniProtKB/Swiss-Prot page.
ORGANISM: The scientific and common names of the species extracted from UniProtKB/Swiss-Prot.
ONTOLOGIES: Any Gene Ontology (GO) terms listed for a protein in UniProtKB/Swiss-Prot.
ISOFORMS: Sequence information for the different isoforms of a protein produced by alternative promoter usage and alternative splicing, if applicable, as annotated in UniProtKB/Swiss-Prot.
MODIFICATIONS: Amino acid residues of a protein that are modified, their position in the sequence, the type of modification, the enzymes that catalyse the modification when known, and publications describing the modification (extracted from phospho.ELM and phosphoSitePlus).
MUTAGENESIS: Amino acid residues of a protein for which experimental mutagenesis data is available, with a description of any functional consequences of the mutation and publications describing the mutation (extracted from UniProtKB/Swiss-Prot).
SECONDARY STRUCTURE: When available, secondary structure of a protein is extracted from UniProtKB/Swiss-Prot, which is assigned by DSSP (Define Secondary Structure of Proteins) to Protein Data Bank entries, i.e. not predicted. Therefor, absence of secondary structure information does not imply the protein is disordered.
3D STRUCTURE: The Protein Data Bank (PDB) identifier of the 3D structure of a protein (e.g. 1AIE), if applicable, and a link to the PDB page describing the structure.
SHORT LINEAR MOTIFS: For motif instances annotated in the Eukaryotic Linear Motif resource (ELM), the ELM class (e.g. LIG_WW_Pin1_4) of the motif and its position in the protein sequence is extracted, and a link to the motif class page in ELM is provided. Motifs belonging to a class not yet annotated in ELM are linked to the Candidates page, which contains a list of candidate motifs that are being annotated, or will be so in the future.
DOMAINS: For domains of a protein annotated in the Protein Families database (Pfam), the Pfam identifier (e.g. PF08563), domain name (e.g. P53_TAD) and its position in the sequence were extracted, and a link to the corresponding page in Pfam is provided. When a domain is not described in Pfam, InterPro accessions and names are used.
DISORDER SCORE: The disorder scores for protein regions are calculated using IUPred, a freely available prediction tool for intrinsically unstructured protein regions. Protein regions with IUPred scores equal to or above 0.4 are considered to be disordered.
Small molecule data
IDENTIFIERS: Small molecules are identified by their Chemical Entities of Biological Interest (ChEBI) name (e.g. 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate), identifier (e.g. CHEBI:18348) and synonyms (e.g. PIP2).
Nucleic acid data
IDENTIFIERS: Nucleic acids are identified by their European Nucleotide Archive (ENA) identifier (e.g. D31852) and description (e.g. Schizosaccharomyces pombe gene for meiRNA, complete sequence).
ORGANISM: The scientific and common names of the species extracted from ENA.
Molecular complexes
IDENTIFIERS: If one binary interaction results in formation of a protein complex that is involved in a second binary interaction, this complex is identified by a switches.ELM-specific identifier and name, i.e. the ID of the interaction that describes the formation of this complex, and a name consisting of the UniProtKB entry names of the two subunits of the complex in alphabetical order connected by a dash (e.g. 1433E_HUMAN-1433E_HUMAN).
Interaction data
IDENTIFIERS: Each interaction is identified by a unique switches.ELM-specific ID. If an interaction results in the formation of a complex that participates in a subsequent interaction, this complex is also attributed a name, consisting of the UniProtKB entry names of the two subunits of the complex in alphabetical order connected by a dash.
PARTICIPANTS: Each interaction is represented as binary, having two molecules participating. Each of the participants is linked to an external resource describing the molecule. If the participant is a protein, binding site name and position are given, and links to external resources describing the interaction sites are provided. If the position of the binding region as given by the authors in a publication is different from that found in the available resources, the positions that map this region to the canonical sequence in UniProtKB were chosen.
SUBCELLULAR LOCALISATION: The subcellular compartment where an interaction occurs is specified by GO cellular component terms.
TEMPORAL DATA: Temporal information for an interaction, for instance during which phase of the cell cycle it occurs, is specified by GO biological process terms for these different phases.
SEQUENTIAL INTERACTIONS: If multiple interactions occur in a specific, well-defined order, they are numbered according to their position in the sequence of binding events.
MUTUALLY EXCLUSIVE INTERACTIONS: If an interaction is mutually exclusive with one or more other interactions, these of these interactions are listed and annotated as mutually exclusive interactions.
REQUIRED INTERACTIONS: If a previous interaction is required to occur for a next interaction to be able to occur, the previous interaction is annotated as a required interaction.
ADDITIONAL INFORMATION: If available, affinity values for interactions are given, in micromoles. Also, some additional notes or remarks concerning an interaction might be annotated.
BIBLIOGRAPHY: Publications describing an interaction are listed using their PubMed identifiers (PMID), which are linked to their abstracts in PubMed.
Regulatory data
IDENTIFIERS: Each switch that is curated in the database is identified by a unique switches.ELM-specific ID and name. The latter consists of the name of the binding site(s) whose functionality is switched and the switch subtype.
SWITCH TYPE - SUBTYPE: Each switch is described by its type and, if applicable, its subtype.
SWITCH MECHANISM: Each switch consists of at least one interaction, which is regulated by switching the function of one of the participants. The switch mechanism describes how the function of one of the interactors is switched to regulate a specific interaction. This either involves the addition of a PTM or the binding of an effector. When the switching mechanism is PTM-dependent, the modified residue, its position in the sequence and the type of modification, using the PSI-MI controlled vocabulary for protein modifications, are annotated. When the switching mechanism is mediated by binding of an effector, the effector is annotated and linked to an external resource for full description.
AFFECTED INTERACTOR: If the switching mechanism depends on PTM or on binding of an effector, the affected interactor specifies to which interactor the modification is added or to which interactor the effector binds. Also, if an interaction describes a modification event, for instance phosphorylation of a protein by a kinase, the affected interactor specifies which interactor is modified.
SWITCH OUTCOME: The switch outcome specifies whether the switching mechanism either induces or inhibits a specific interaction.
TEMPORAL DATA: Temporal information for a switch, for instance during which phase of the cell cycle it occurs, is specified by GO biological process terms for these different phases.
PATHWAYS: A list of pathways in which a switch is involved is provided, with links to external resources describing these pathways.
DESCRIPTION: A text description of the switch and the context of its regulatory role is provided.
ADDITIONAL INFORMATION: When applicable, figures in specific publications that provide detailed graphical information about the regulatory mechanism that is described and any additional notes about a specific switch are mentioned.
BIBLIOGRAPHY: Publications describing regulation of an interaction are listed using their PubMed identifiers (PMID), which are linked to their abstracts in PubMed
Inferred versus curated data
LOCALISATION DATA: The subcellular localisation where a motif-mediated interaction occurs is inferred from the GO cellular component terms annotated for the motif in the Eukaryotic Linear Motif resource (ELM). Experimentally validated subcellular localisation of an interaction is curated from the literature.
TEMPORAL DATA Temporal information for an interaction, for instance during which phase of the cell cycle it occurs, is inferred from the Reactome pathway database. An interaction is inferred to occur in a cell cycle phase that is annotated in Reactome and shared by both participants. The switch is inferred to occur in those phases that are shared by the interactions involved in the switch. Experimentally validated cell cycle phases for an interaction or a switch are curated from the literature.
MODIFICATION DATA: For switches mediated by a specific covalent modification of a residue in an interactor, all enzymes known to catalyse this modification are extracted from UniProtKB/Swiss-Prot and phospho.ELM and are inferred to mediate the switch. Enzymes that were experimentally shown to regulate the switch are manually curated from the literature.
PATHWAY DATA: In which pathway a switch is involved, is inferred from the Reactome pathway database. If at least two participants that are involved in a switch have a common pathway(s) annotated in Reactome, the switch in which these participants are involved, is inferred to be involved in this common pathway(s). Experimentally validated pathways for a switch are curated from the literature.
switches.ELM Downloads and Usage Policy
The data that is curated in switches.ELM is not our own, but was collected from peer-reviewed publications available in PubMed. When using any of these data, please refer to the original corresponding publications to which it is linked. Data downloaded from switches.ELM is only free for academic use, not for commercial use.
All motif-based switch data in switches.ELM can be downloaded in tsv format here:
Download switches as tsv
Download inferred pathway/modification/location data as tsv