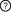
This is a
sequential switch. The interactions in this switch take place in a
defined order. The order is defined by numbers to the left of each interaction. More than one interaction can occur in each step, these interactions occur independently but are necessary for future step