Description: Participants: Interactions 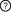 This is a sequential switch. The interactions in this switch take place in a defined order. The order is defined by numbers to the left of each interaction. More than one interaction can occur in each step, these interactions occur independently but are necessary for future step This is a sequential switch. The interactions in this switch take place in a defined order. The order is defined by numbers to the left of each interaction. More than one interaction can occur in each step, these interactions occur independently but are necessary for future step1Interaction #1 Ywhab - Ywhab Interfaces (1) 14-3-3 protein in 14-3-3 protein beta/alpha (Ywhab) (2) 14-3-3 protein (5-238) in 14-3-3 protein beta/alpha (Ywhab) 2Interaction #2 Ksr1 - Interaction #1 Ywhab - Ywhab Requires Interaction #1 Ywhab - Ywhab Interfaces (3) LIG_14-3-3_3 motif (294RSKSHE299) in Kinase suppressor of Ras 1 (Ksr1) (4) Interaction #1 Ywhab - Ywhab Interaction Regulation PTM-dependent Induction (Phosphorylation of S297 on Kinase suppressor of Ras 1 (Ksr1)) of the Kinase suppressor of Ras 1 (Ksr1) LIG_14-3-3_3 motif - 14-3-3 protein interaction Regulatory Enzymes for switch Modifying enzymes for residue: S297: MAP/microtubule affinity-regulating kinase 3 (Mark3) 2Interaction #3 Ksr1 - Interaction #1 Ywhab - Ywhab Requires Interaction #1 Ywhab - Ywhab Interfaces (5) LIG_14-3-3_3 motif (389RTESVP394) in Kinase suppressor of Ras 1 (Ksr1) (6) Interaction #1 Ywhab - Ywhab Interaction Regulation PTM-dependent Induction (Phosphorylation of S392 on Kinase suppressor of Ras 1 (Ksr1)) of the Kinase suppressor of Ras 1 (Ksr1) LIG_14-3-3_3 motif - 14-3-3 protein interaction Regulatory Enzymes for switch Modifying enzymes for residue: S392: MAP/microtubule affinity-regulating kinase 3 (Mark3) Inferred Regulatory Enzymes for switch Putative modifying enzymes for residue: S392 : Nucleoside diphosphate kinase A (NME1), Mitogen-activated protein kinase kinase kinase 7 (Map3k7). Context Switch localisation Curated: cytosol ELM curated: nucleus, cytosol, internal side of plasma membrane Switch pathways Curated: MAPK signaling pathway, PI3K-Akt signaling pathway References (1) Identification of constitutive and ras-inducible phosphorylation sites of KSR: implications for 14-3-3 binding, mitogen-activated protein kinase binding, and KSR overexpression. See also Other switches involving participants |
|
Please send any suggestions/comments to: switches@elm.eu.org

