Description: Participants: Interactions 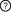 This is a sequential switch. The interactions in this switch take place in a defined order. The order is defined by numbers to the left of each interaction. More than one interaction can occur in each step, these interactions occur independently but are necessary for future step This is a sequential switch. The interactions in this switch take place in a defined order. The order is defined by numbers to the left of each interaction. More than one interaction can occur in each step, these interactions occur independently but are necessary for future step1Interaction #1 rad24 - rad24 Interfaces (1) 14-3-3 protein in DNA damage checkpoint protein rad24 (rad24) (2) 14-3-3 protein (6-241) in DNA damage checkpoint protein rad24 (rad24) 2Interaction #2 mei2 - Interaction #1 rad24 - rad24 Requires Interaction #1 rad24 - rad24 Interfaces (3) LIG_14-3-3_1 motif (435RTESSP440) in Meiosis protein mei2 (mei2) (4) Interaction #1 rad24 - rad24 Interaction Regulation PTM-dependent Induction (Phosphorylation of S438 on Meiosis protein mei2 (mei2)) of the Meiosis protein mei2 (mei2) LIG_14-3-3_1 motif - 14-3-3 protein interaction Regulatory Enzymes for switch Modifying enzymes for residue: S438: Negative regulator of sexual conjugation and meiosis (ran1) 2Interaction #3 mei2 - Interaction #1 rad24 - rad24 Requires Interaction #1 rad24 - rad24 Interfaces (5) LIG_14-3-3_2 motif (523RRSLTVG529) in Meiosis protein mei2 (mei2) (6) Interaction #1 rad24 - rad24 Interaction Regulation PTM-dependent Induction (Phosphorylation of T527 on Meiosis protein mei2 (mei2)) of the Meiosis protein mei2 (mei2) LIG_14-3-3_2 motif - 14-3-3 protein interaction Regulatory Enzymes for switch Modifying enzymes for residue: T527: Negative regulator of sexual conjugation and meiosis (ran1) Context References (1) 14-3-3 protein interferes with the binding of RNA to the phosphorylated form of fission yeast meiotic regulator Mei2p. See also Other switches involving participants |
|
Please send any suggestions/comments to: switches@elm.eu.org

